In today’s AI and data science applications, Python has become one of the most dominant programming languages. To help users use JuiceFS more efficiently in these scenarios, we’ve introduced the JuiceFS Python SDK in the Community Edition 1.3. This SDK not only simplifies access to JuiceFS but also improves usability in constrained environments.
For example, in serverless scenarios where users often cannot mount a file system, the Python SDK enables direct read/write operations on JuiceFS without mounting. This significantly improves flexibility. Moreover, in high-performance use cases, the Python SDK delivers superior performance and experience.
This article will briefly cover the Python SDK’s features and our performance optimization practices. We welcome community users to try this new feature and share feedback.
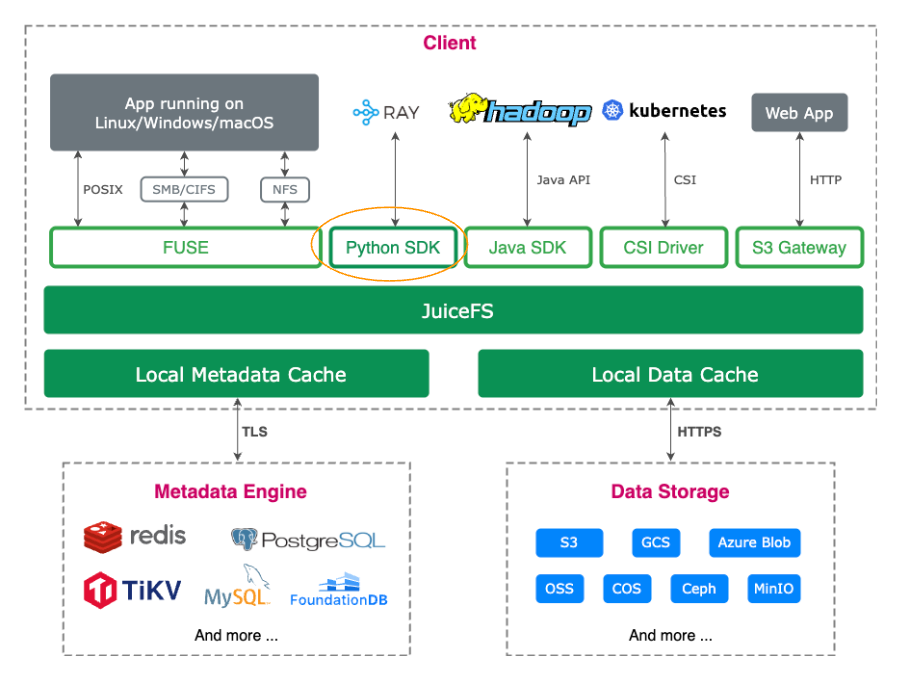
Python SDK overview
The JuiceFS Python SDK provides multiple interfaces, covering diverse use cases—from basic file operations to ecosystem integrations.
The JuiceFS client interface
juicefs.Client encapsulates critical functionalities of the JuiceFS client. It supports standard file system operations (such as open and rename) and extended features like warmup, summary, and rmr, enabling developers to implement complex management tasks.
Users can quickly explore documentation via Python’s help() function, viewing methods and usage for each class.
Initializing the JuiceFS client is easy—just provide the file system name (name) and metadata address (meta). Once initialized, developers can interact with JuiceFS as if it were a local file system (for example, listing root directories or modifying files).
Native Python file interface compatibility
The file operation interfaces provided by the JuiceFS Python SDK maintain full compatibility with file objects returned by Python's native open() method. In other words, when a user opens a file using client.open(), the returned object supports standard Python file operations such as read(), write(), seek(), and close().
This allows users to seamlessly integrate the JuiceFS file system into their existing Python code logic without needing to learn or adapt to new APIs. Whether for data preprocessing, model training, or log management scenarios, users can perform file read/write operations using the Python SDK.
fsspec support for seamless Ray integration
To better support high-performance data processing in AI training and data science scenarios, the JuiceFS Python SDK provides native fsspec interface support. As a standardized abstraction layer in Python’s ecosystem, fsspec unifies file system access and is widely adopted by mainstream AI frameworks like Ray.
JuiceFS seamlessly integrates with AI toolchains, enabling users to:
- Access JuiceFS storage as easily as local disks
- Maintain existing application logic without code modifications
- Enhance I/O performance while reducing management/scale costs
- Below is an example demonstrating how to configure JuiceFS as an
fsspecbackend file system:
import fsspec
import ray
import sys
sys.path.append('.')
import sdk.python.juicefs.juicefs.spec
jfs = fsspec.filesystem("jfs", auto_mkdir=True, name="myjfs", meta="redis://localhost")
dsjfs = ray.data.read_csv('/ray_demo_data.csv', filesystem=jfs)
dsjfs.count()
Only the essential parameters (file system name and metadata address) need to be specified. All other operations remain consistent with standard fsspec-compatible file systems.
This design significantly enhances JuiceFS' usability in distributed computing frameworks like Ray, streamlining integration with existing data loading and processing pipelines.
Extended APIs
In addition to standard file operations, the JuiceFS Python SDK provides a set of extended APIs such as summary and info—interfaces already familiar to many users from the command-line tools.
In the Python SDK, these extended commands return results in dictionary (dict) format, enabling easy access and indexing within scripts. For example, when using the summary interface, you can directly retrieve file or directory statistics via key-value pairs:
summary_info = client.summary("/path/to/dir")
print(summary_info["fileCount"])
This dictionary-structured return format significantly enhances automation scripting efficiency and simplifies integration with existing Python data processing workflows.
Performance optimization: FFRecord data loading with 3x speedup over FUSE
When accessing data via FUSE, the upper limit of a single I/O request is 128 KB. Even if it can be increased to 1 MB through direct I/O, a complete data read is divided into multiple small blocks by the kernel. This results in additional I/O request amplification. These small requests are submitted synchronously and serially. This will significantly increase the latency and reduce the overall throughput performance when accessing cold data (that is, data that has not been cached locally). At the same time, multiple consecutive requests also start the JuiceFS readahead process, resulting in unnecessary read amplification.
To bypass FUSE's request size limitations, our Python SDK enables larger I/O requests to mitigate fragmentation-related performance degradation. To validate the Python SDK's performance, we developed an FFRecord dataloader demo. FFRecord is an open-source data format designed to:
- Consolidate small files
- Reduce read overhead
- Enable random batch reading
- Provide data verification
When loading FFRecord data through FUSE, a critical limitation is the constrained request size: FUSE defaults to a maximum 128 KB read request. This forces large reads to be fragmented into 128 KB chunks, significantly amplifying I/O latency. In addition, when processing sequential reads, the JuiceFS client automatically performs readahead operations within a certain range. This will cause 2x~4x read amplification, further increasing the system burden.
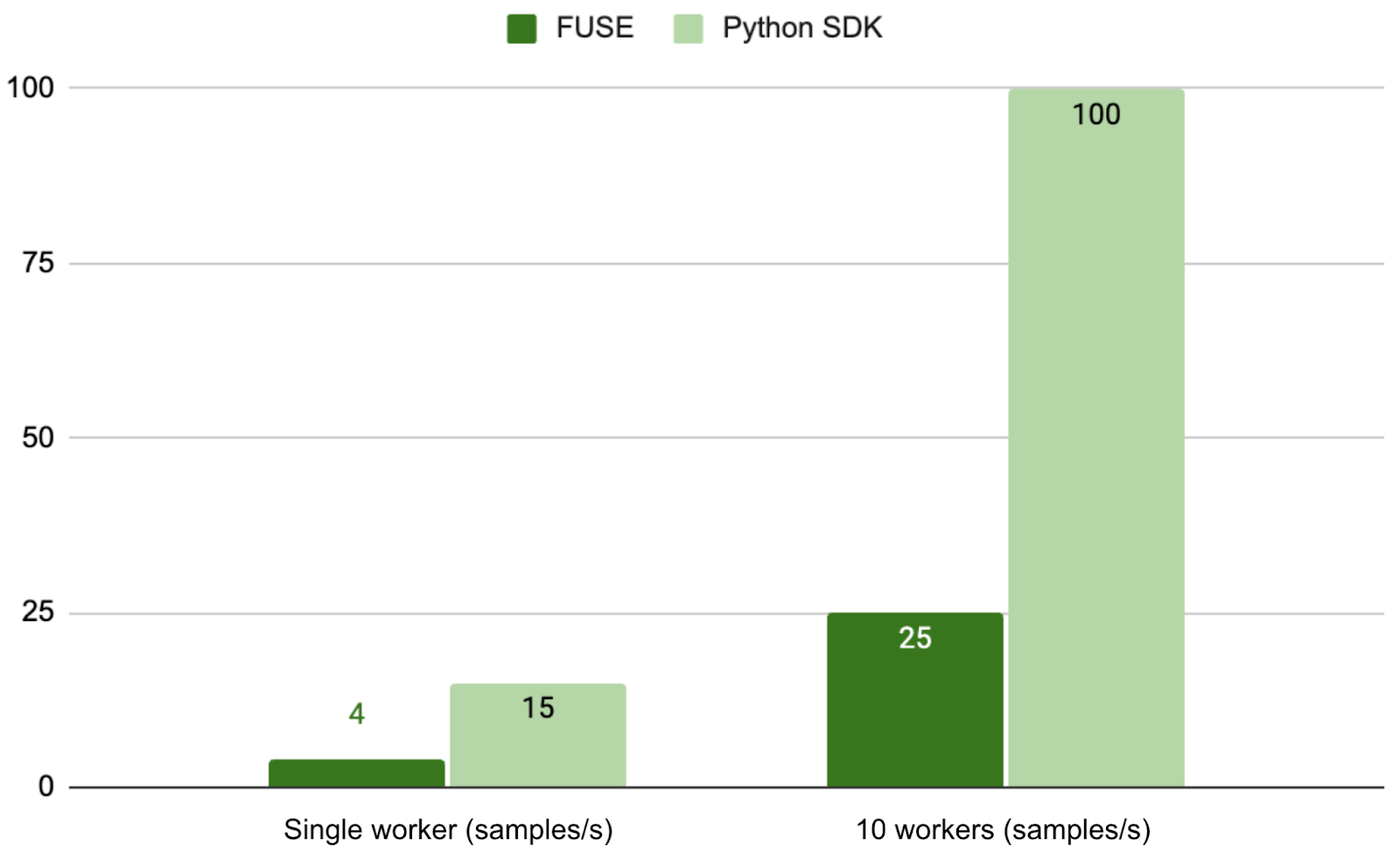
To address this, we implemented an optimized FFRecord dataloader using the Python SDK. Our benchmark tests used a dataset containing 1,000 samples (each approximately 3 MB ± 500 KB). The test results demonstrate 3.75× and 4× performance improvements in single-worker and multi-worker scenarios respectively when using the Python SDK versus conventional FUSE mounting.
Implementation logic of the demo:
At the lower level, the file_reader class parses and validates the FFRecord file format, handling both samples and file headers. During initialization, the header is loaded to record the offset information of all samples in the specified format, supporting subsequent read operations. In addition, file_reader provides two interfaces—read_one and read_batch—enabling flexible calls for either single-sample or batch-sample reading, while also facilitating future development and extensions.
Current limitations in the Python SDK implementation:
The current implementation has certain constraints, such as lacking support for concurrent read_batch execution (only serial processing is available). Future updates will move concurrent read_batch operations to the dynamic library layer or implement asynchronous logic in Python. Furthermore, when enabling concurrent loading with num_workers, each worker process must initialize its own JuiceFS client instance. This is because client resources cannot be shared when the main process forks child processes, requiring independent initialization. While this design introduces some overhead, the overall impact remains minimal.
Higher-level implementation:
The dataset and dataloader implementations at the upper level are simple—they simply invoke the underlying reader to complete the loading process. We have also included a dataset generation script to produce demo.ffr files for testing and loading. All related code has been organized in the community edition repository for reference.
Python SDK installation
The JuiceFS Python SDK is now available in v1.3-beta1. This SDK reuses the underlying dynamic library from the JuiceFS Java SDK, which implements the core objects and functionalities of the JuiceFS client.
In the Python SDK, we expose these underlying capabilities through Python interfaces, allowing users to directly invoke them in Python environments. As a result, the installation process is straightforward, consisting of just two key steps:
- Compile the dynamic library
- Package and install the Python module
For detailed instructions, see the Python SDK document.
Summary
The Python SDK brings more flexible integration for JuiceFS, offering:
- Compatibility with Python's native file interfaces
- Support for the
fsspecabstraction layer (enabling seamless use with AI components like Ray)
In terms of high-performance access, we explored the scenario of FFRecord data loading and verified its performance potential.
The complete example code is available in the community repository. We welcome all users to test, provide feedback, and collaborate on further enhancing the SDK's capabilities. If you have any questions for this article, feel free to join JuiceFS discussions on GitHub and community on Slack.















